An Overlooked Source of (Massive) Run-time Optimization in KMeans
Another issue with KMeans and here's how to address it.
Yesterday, we learned how the mini-batch implementation of the KMeans clustering algorithm works.
More specifically, we discussed how during the centroid reassignment step in traditional KMeans, all data points must be available in memory to average over.
But in large datasets, keeping all data points in memory can be difficult.
MiniBatchKMeans is a mini-batch implementation of KMeans, which addresses this issue by maintaining the following details for every centroid while iterating through all mini-batches:
A “
sum-vector” to store the sum of data points vectors assigned to the centroidA “
count” variable to store the number of data points vectors assigned to the centroid.
After iterating through all mini-batches, we perform a scalar division between sum-vector and count to get the average vector:
This average vector will precisely tell us the new location of the centroid as if all data points were present in memory.
This solves the memory bottleneck issues of KMeans.
In fact, a mini-batch approach can also offer a run-time improvement because, while typically, vectorization provides magical run-time improvements when we have a bunch of data, the performance often degrades after a certain point.

Thus, by loading fewer training instances at a time into memory, we can get a better training run-time.
For more details, please refer to yesterday’s issue.
Another limitation of KMeans
Nevertheless, there’s another major limitation of both KMeans and MiniBatchKMeans, which we didn’t discuss yesterday.
To recall, KMeans is trained as follows:
Step 1) Initialize centroids
Step 2) Find the nearest centroid for each point
Step 3) Reassign centroids
Step 4) Repeat until convergence
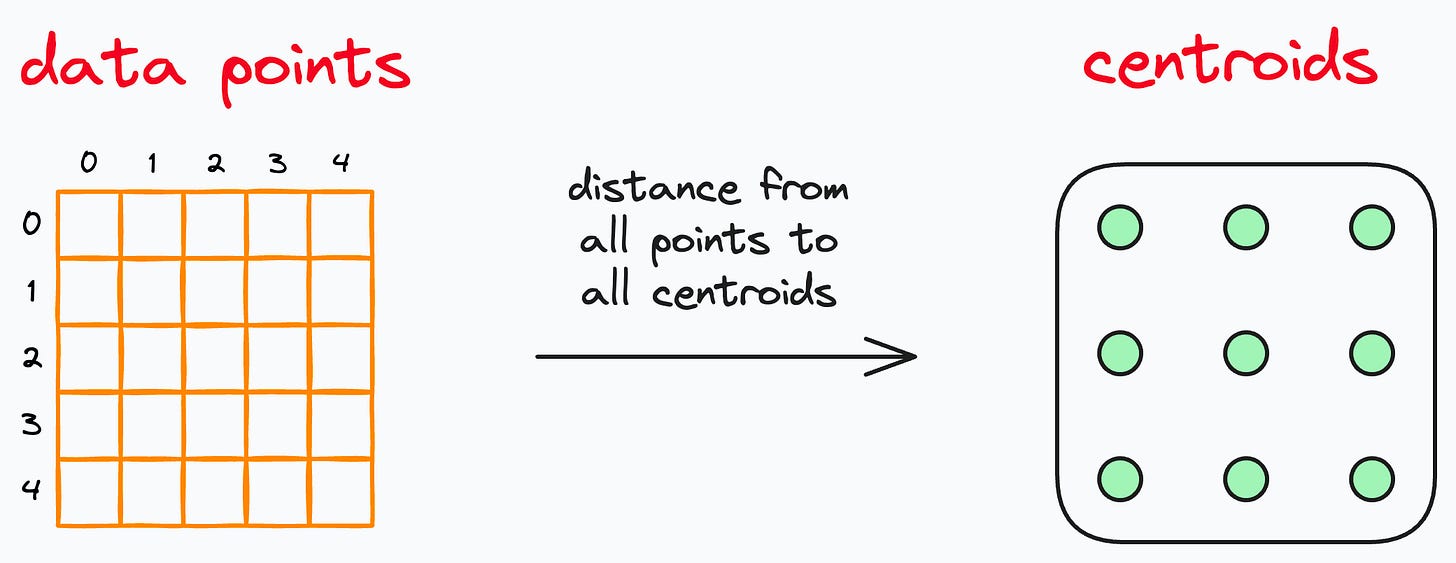
But in this implementation, even “Step 2” has a run-time bottleneck, as this step involves a brute-force and exhaustive search.
In other words, it finds the distance of every data point from every centroid.
As a result, this step isn’t optimized, and it takes plenty of time to train and predict.
This is especially challenging with large datasets.
To speed up KMeans, one of the implementations I usually prefer, especially on large datasets, is Faiss by Facebook AI Research.
To elaborate further, Faiss provides a much faster nearest-neighbor search using approximate nearest-neighbor search algorithms.
It uses “Inverted Index”, which is an optimized data structure to store and index the data point.
This makes performing clustering extremely efficient, especially on large datasets, which is also evident from the image below:
As shown above, on a dataset of 500k data points (1024 dimensions), Faiss is roughly 20x faster than KMeans from Sklearn, which is an insane speedup.
What’s more, Faiss can also run on a GPU, which can further speed up your clustering run-time performance.
👉 Get started with Faiss here: GitHub.
👉 Over to you: What are some other limitations of the KMeans algorithm?
👉 If you liked this post, don’t forget to leave a like ❤️. It helps more people discover this newsletter on Substack and tells me that you appreciate reading these daily insights.
The button is located towards the bottom of this email.
Thanks for reading!
Latest full articles
If you’re not a full subscriber, here’s what you missed last month:
DBSCAN++: The Faster and Scalable Alternative to DBSCAN Clustering
Federated Learning: A Critical Step Towards Privacy-Preserving Machine Learning
You Cannot Build Large Data Projects Until You Learn Data Version Control!
Sklearn Models are Not Deployment Friendly! Supercharge Them With Tensor Computations.
Deploy, Version Control, and Manage ML Models Right From Your Jupyter Notebook with Modelbit
Gaussian Mixture Models (GMMs): The Flexible Twin of KMeans.
To receive all full articles and support the Daily Dose of Data Science, consider subscribing:
👉 Tell the world what makes this newsletter special for you by leaving a review here :)
👉 If you love reading this newsletter, feel free to share it with friends!