Breathing KMeans vs KMeans
Robustify KMeans with centroid addition and removal.
KMeans is widely used for its simplicity and effectiveness as a clustering algorithm.
Yet, we all know that its performance is entirely dependent on the centroid initialization step.
Thus, it is likely that we may obtain inaccurate clusters, as depicted below:
Of course, rerunning with different initialization does help at times.
But I have never liked the unnecessary run-time overhead it introduces.
So today, let me share a neat and supercharged upgrade to KMeans, which addresses this issue while also producing better clustering results.
It’s called the Breathing KMeans algorithm.
Let’s understand how it works.
Step 1: Run Kmeans
First, we run the usual KMeans clustering only once, i.e., without rerunning the algorithm with a different initialization.
This gives us the location of “k” centroids, which may or may not be accurate.
Step 2: Breathe in step
To the “k” centroids obtained from Step 1, we add “m” new centroids.
As per the research paper of Breathing Kmeans,
m=5was found to be good value.
Now, you might be thinking, where do we add these “m” centroids?
The addition of new centroids is decided based on the error associated with a centroid.
Simply put, a centroid’s error is the sum of the squared distance to the points associated with that centroid.
Thus, we add “m” centroids in the vicinity of centroids with high error.
Let’s understand more intuitively why this makes sense.
In the above KMeans clustering results:
The centroid at the top has a high error.
All other centroids have relatively low error.
Intuitively speaking, if a centroid has a very high error, it is possible that multiple clusters are associated with it.
Thus, we would want to split this cluster.
Adding new centroids near clusters with high error will precisely fulfill this objective.
After adding “m” new centroids, we get a total of “k+m” centroids.
Finally, we run KMeans again with “k+m” centroids only once.
This gives us the location of “k+m” centroids.
Step 3: Breathe out step
Next, we want to remove “m” centroids from the “k+m” centroids obtained above.
Here, you might be thinking, which “m” centroids should we remove?
The removal of centroids is decided based on the “utility” of a centroid.
Simply put, a centroid’s utility is proportional to its distance from all other centroids.
The greater the distance, the more isolated it will be.
Hence, the more the utility.
This makes intuitive sense as well.
If two centroids are pretty close, they will have low utility.
Thus, they are likely in the same cluster, and we must remove one of them.
This is demonstrated below:
After removing one of the low-utility centroids, the other centroid becomes very useful.
So, in practice, after removing one centroid, we update the utility values of all other centroids.
We repeat the process until all “m” low-utility centroids have been removed.
This gives back “k” centroids.
Finally, we run KMeans with these “k” centroids only once.
Step 4: Decrease m by 1.
Step 5: Repeat Steps 2 to 4 until m=0.
Done!
These repeated breathing cycles (breathe-in and breathe-out steps) almost always provide a faster and better solution than standard KMeans with repetitions.
In each cycle:
New centroids are added at “good” locations. This helps in splitting clusters occupied by a single centroid.
Low-utility centroids are removed. This helps in eliminating centroids that are likely in the same cluster.
As a result, it is expected to converge to the optimal solution faster.
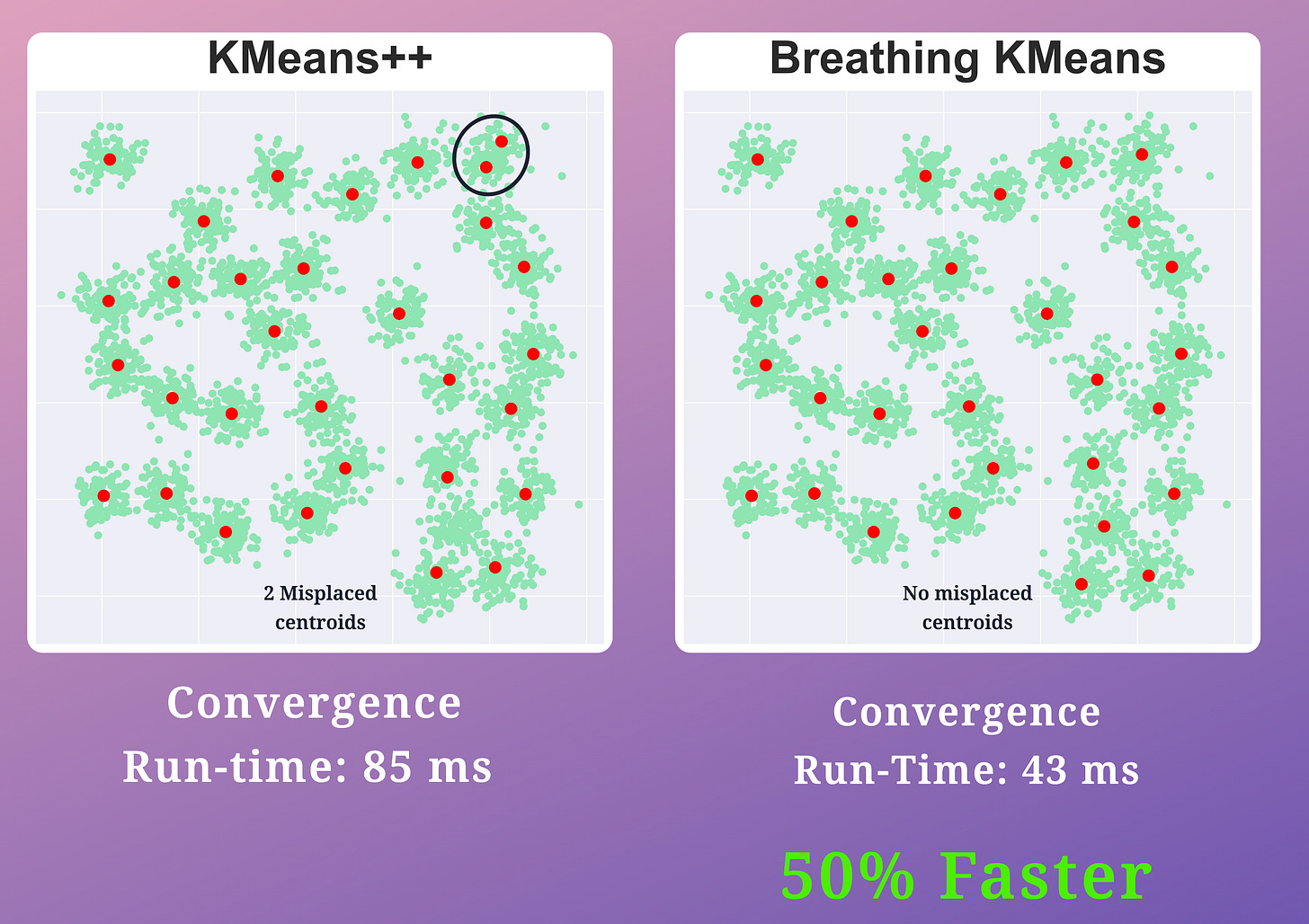
The effectiveness of Breathing KMeans over KMeans is evident from the image below:
KMeans produced two misplaced centroids
Breathing KMeans accurately clustered the data with a 50% run-time improvement.
There is also an open-source implementation of Breathing KMeans, with a sklearn-like API.
To get started, install the bkmeans library:
pip install bkmeansNext, run the algorithm as follows:
from bkmeans import BKMeans
bkm = BKMeans(n_clusters = 100)
bkm.fit(X)Isn’t that a cool upgrade to KMeans?
That said, data conformity is another big issue with KMeans, which makes it highly inapplicable in many data situations.
These two guides cover distribution-based and density-based clustering, which address KMeans’ limitations in specific data situations:
👉 Over to you: What are some other ways to improve KMeans’ clustering and its run-time?
Thanks for reading!
Whenever you are ready, here’s one more way I can help you:
Every week, I publish 1-2 in-depth deep dives (typically 20+ mins long). Here are some of the latest ones that you will surely like:
[FREE] A Beginner-friendly and Comprehensive Deep Dive on Vector Databases.
A Detailed and Beginner-Friendly Introduction to PyTorch Lightning: The Supercharged PyTorch
You Are Probably Building Inconsistent Classification Models Without Even Realizing
Why Sklearn’s Logistic Regression Has no Learning Rate Hyperparameter?
PyTorch Models Are Not Deployment-Friendly! Supercharge Them With TorchScript.
Federated Learning: A Critical Step Towards Privacy-Preserving Machine Learning.
You Cannot Build Large Data Projects Until You Learn Data Version Control!
To receive all full articles and support the Daily Dose of Data Science, consider subscribing:
👉 If you love reading this newsletter, feel free to share it with friends!
👉 Tell the world what makes this newsletter special for you by leaving a review here :)