Breathing KMeans vs KMeans
Robustify KMeans with centroid addition and removal.
Integrate coding Agents into your workflows
Using the Codegen SDK, you can now programmatically interact with your AI Coding Agents:
You can use it to:
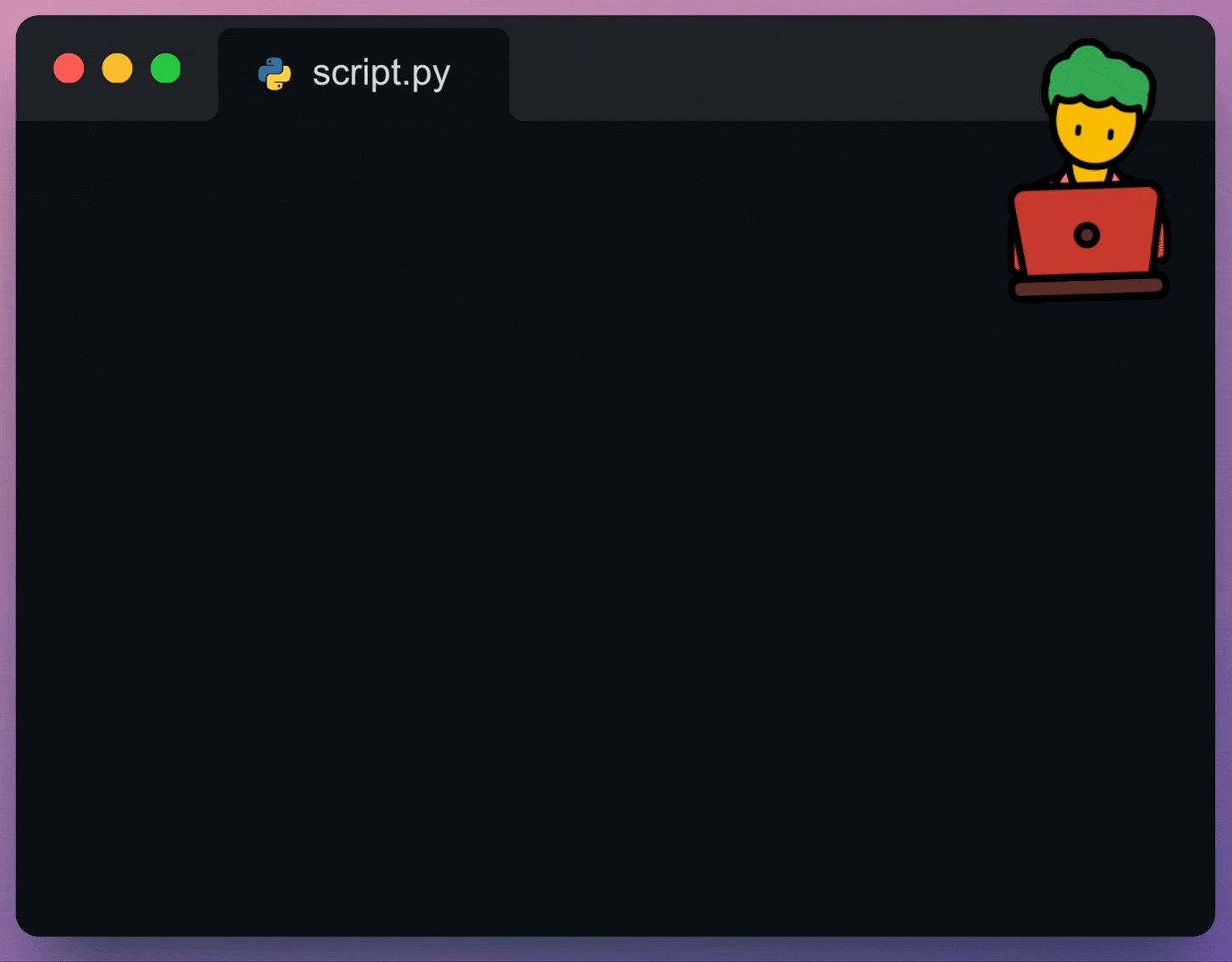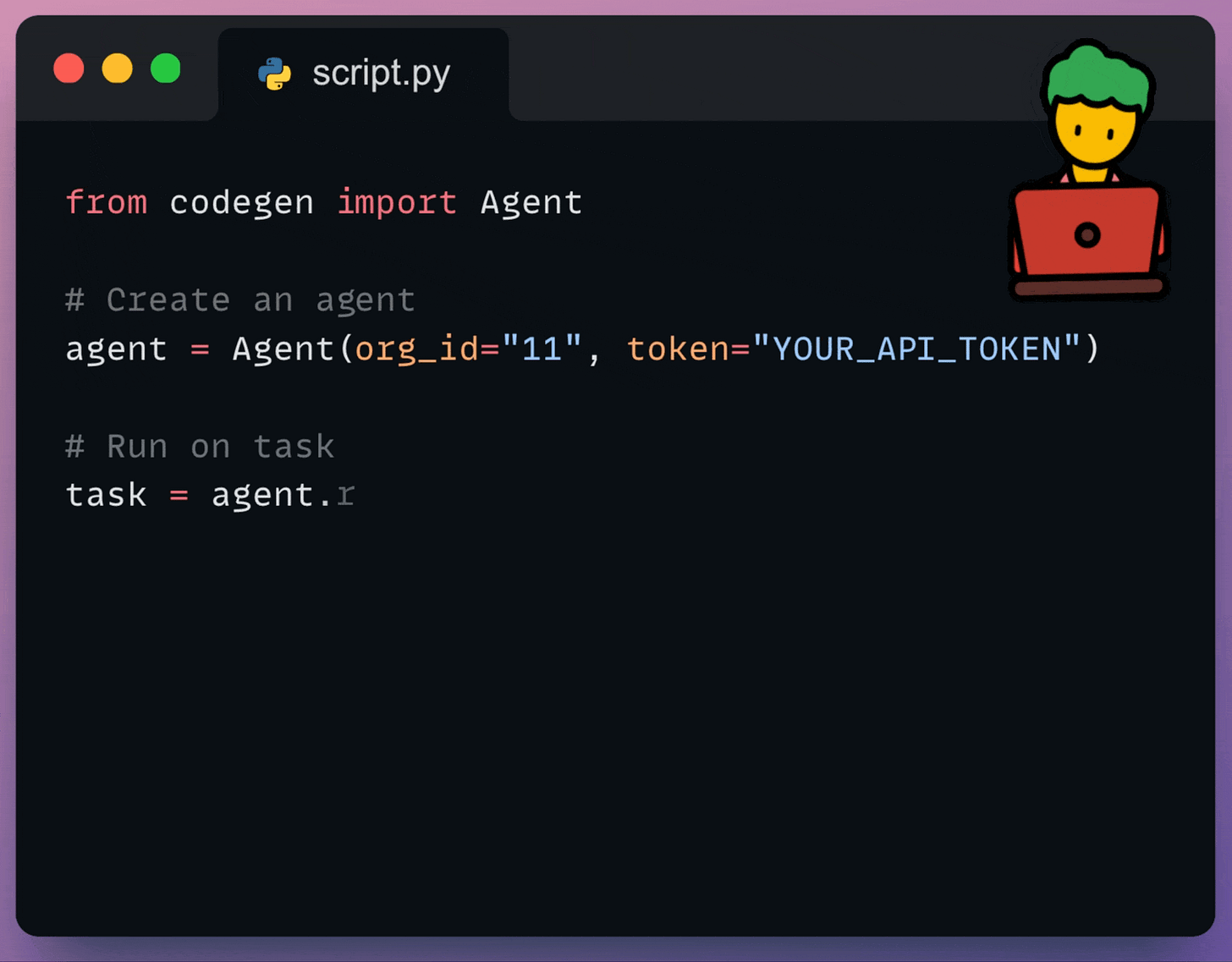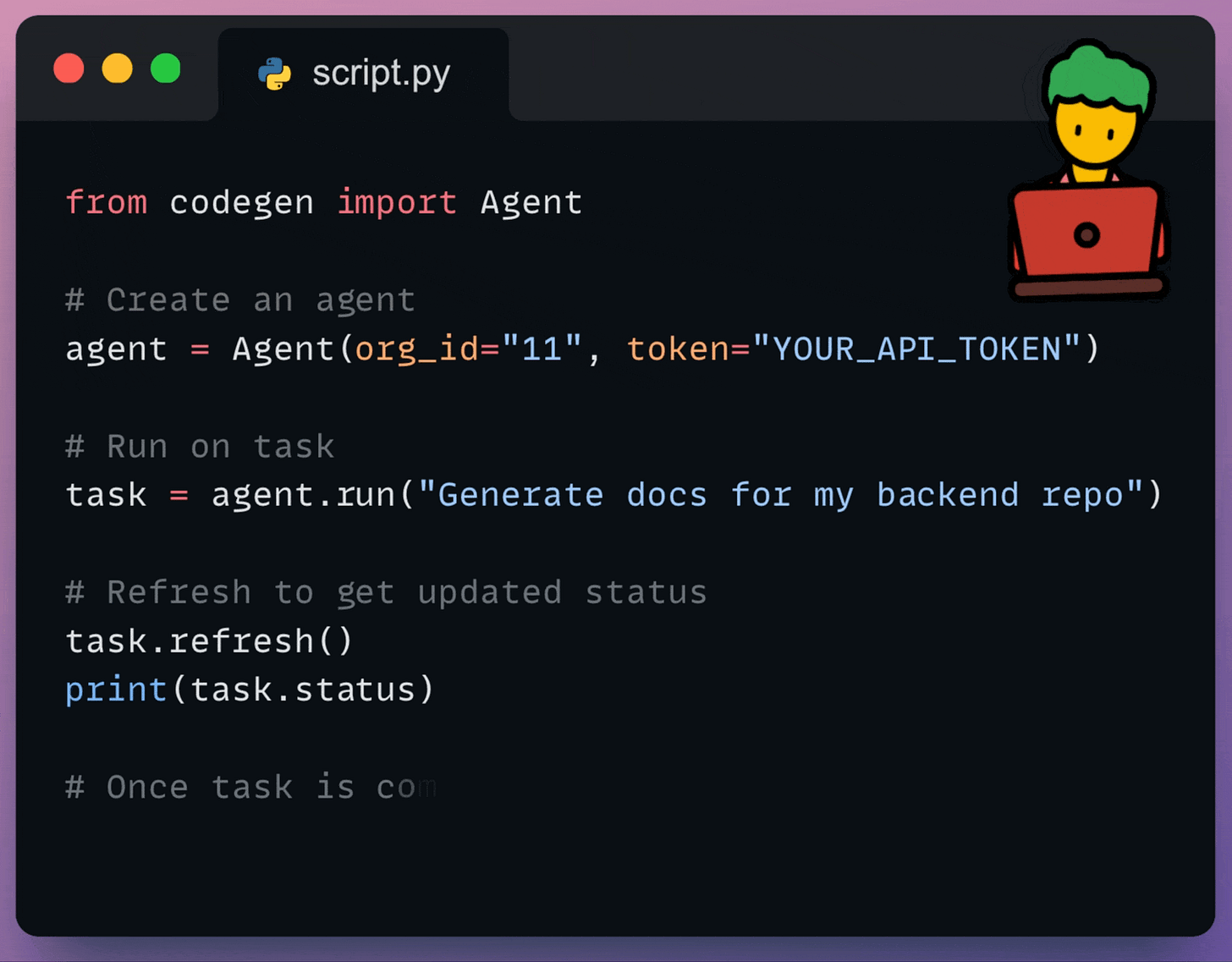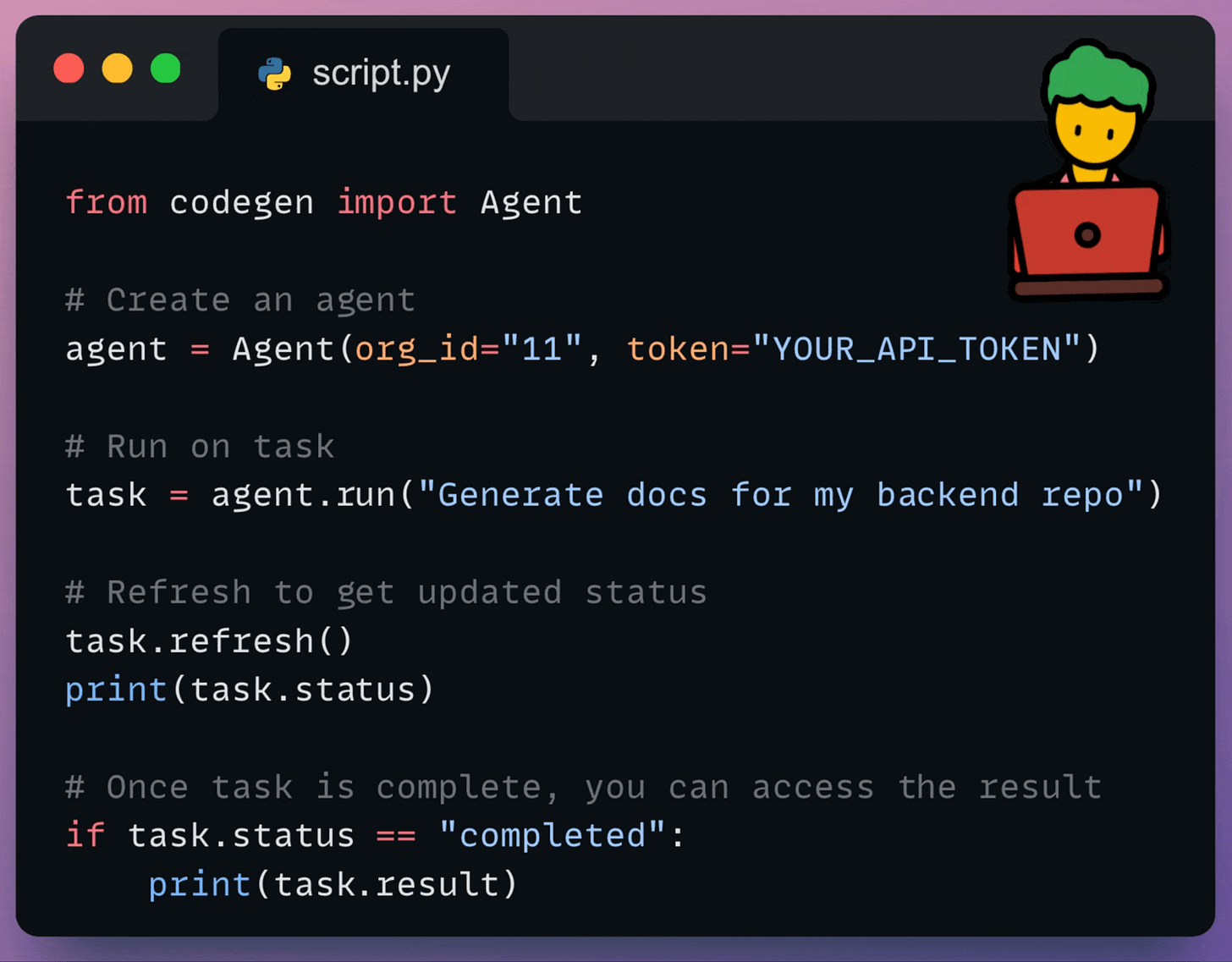
Assign tasks like implementing features, fixing bugs, writing tests, or improving documentation to the agent.
Trigger agent tasks from your CI/CD pipelines, scripts, or other development tools.
Supply the agent with specific instructions, relevant code snippets, or background information to ensure it performs tasks according to your requirements.
Essentially, the SDK allows you to leverage Codegen’s AI capabilities wherever you can run Python code.
Here’s the documentation to learn more →
Breathing KMeans vs KMeans
Since KMeans’ performance heavily depends on the centroid initialization step, it is always advised to run the algorithm multiple times with different initializations.
But this repetition introduces an unnecessary run-time overhead.
The Breathing KMeans algorithm solves this issue while providing better clustering results than KMeans.
There is also an open-source implementation of Breathing KMeans with a sklearn-like API.
To get started, install the bkmeans library and run the algorithm as follows:
Done!
If you are curious, we have covered how Breathing KMeans works in the next section.
On a side note, data conformity is another big issue with KMeans, which makes it highly inapplicable in many data situations.
These three guides cover distribution-based and density-based clustering, which address KMeans’ limitations in specific data situations:
Step 1: Run Kmeans
First, run the usual KMeans clustering only once, i.e., without rerunning it with a different initialization.
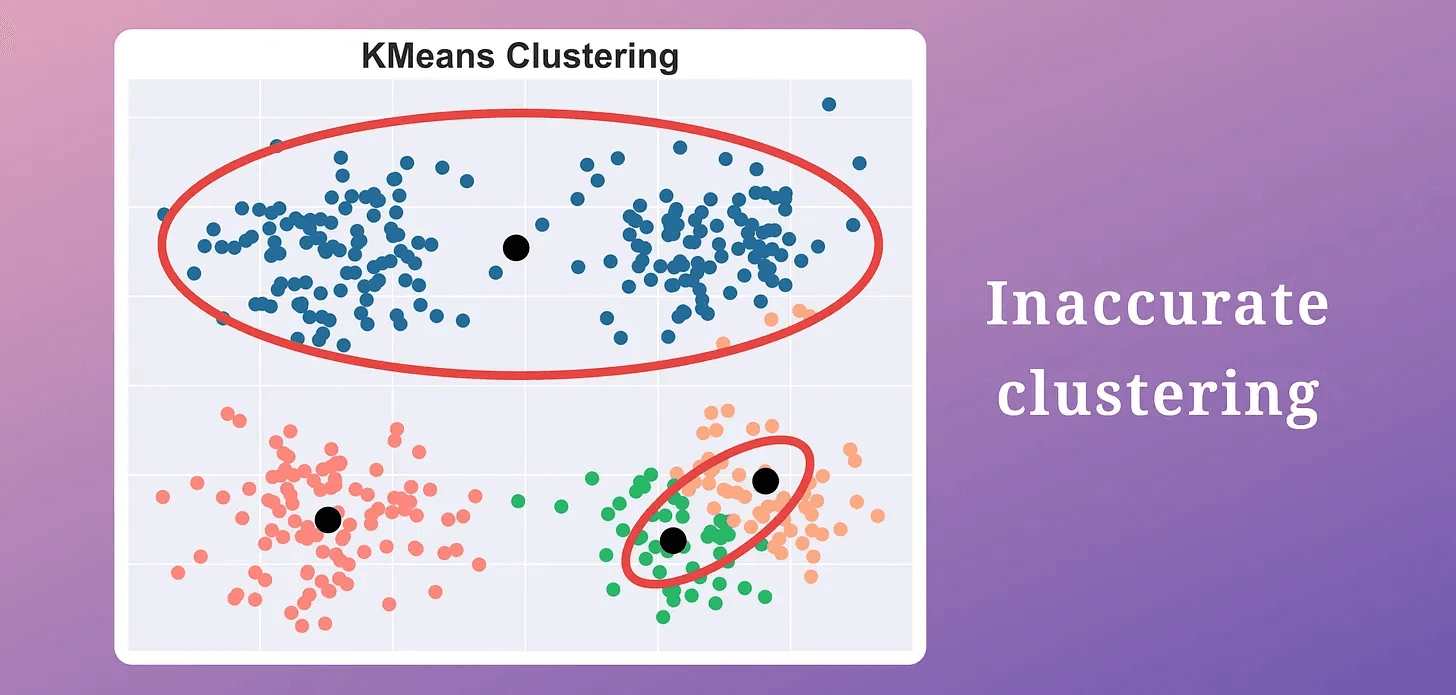
This gives us the location of “k” centroids, which may be inaccurate.
Step 2: Breathe in step
Add “m” new centroids to the “k” centroids obtained above (usually m=5).
Where to place them?
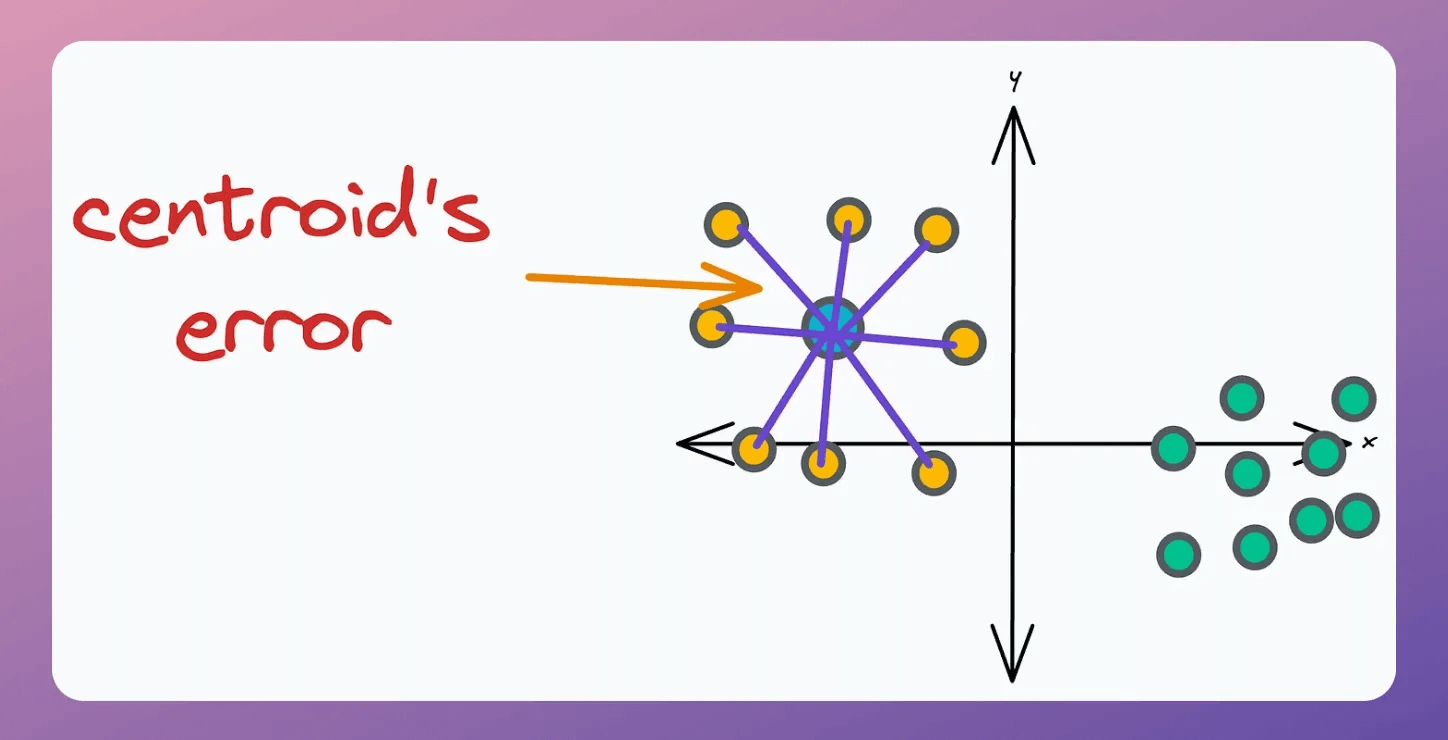
This is decided based on the error associated with the “k” existing centroids. A centroid’s error is the sum of the squared distance to its associated points.
Thus, we add “m” centroids near centroids with high error.
To understand this intuitively, consider these clustering results:
The centroid at the top has a high error.
All other centroids have relatively low error.
Intuitively speaking, if a centroid has a very high error, multiple clusters may be associated with it.
Thus, we must split this cluster by adding new centroids near centroids with high error.
This gives us a total of “k+m” centroids.
Next, rerun KMeans with “k+m” centroids only once.
Step 3: Breathe out step
Next, we should remove “m” centroids from the “k+m” centroids obtained above.
Which “m” centroids should we remove?
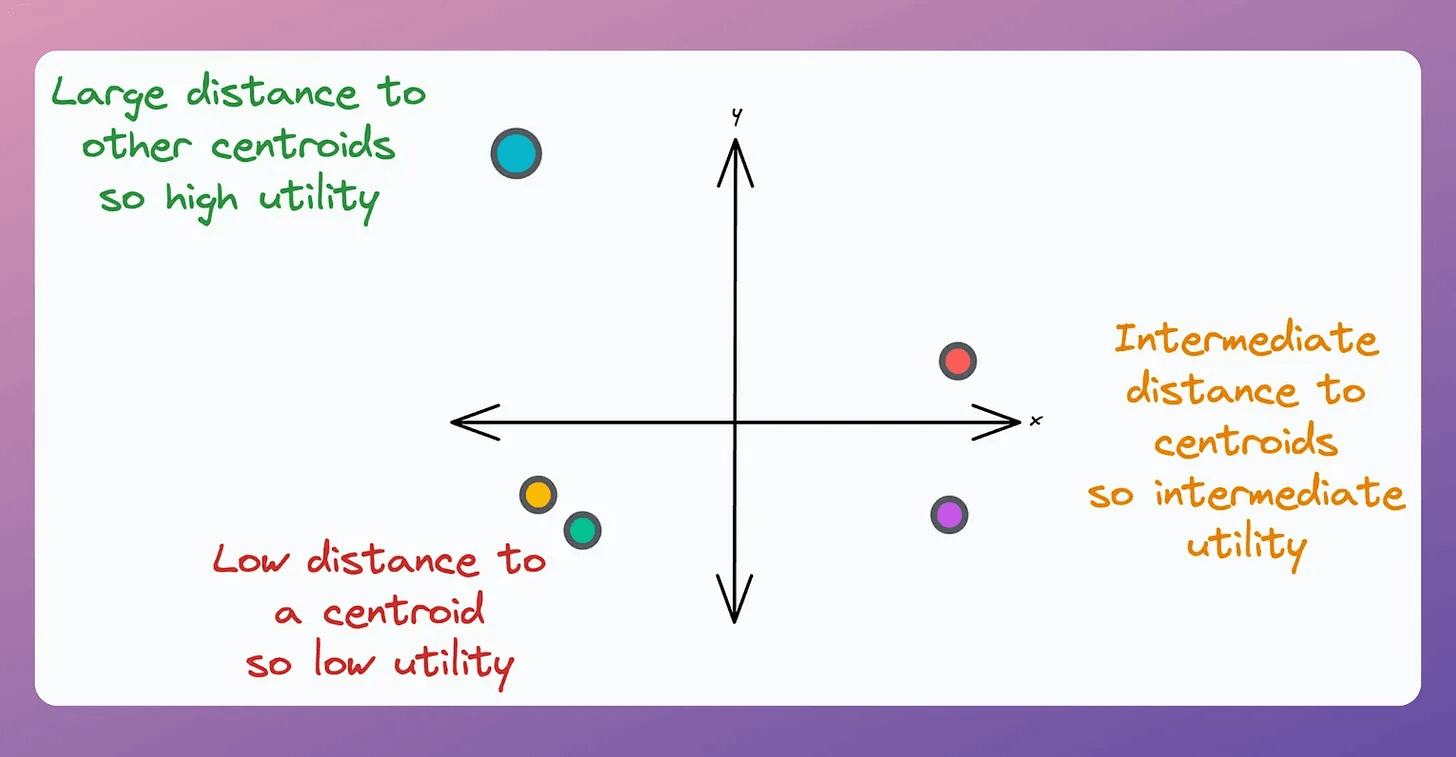
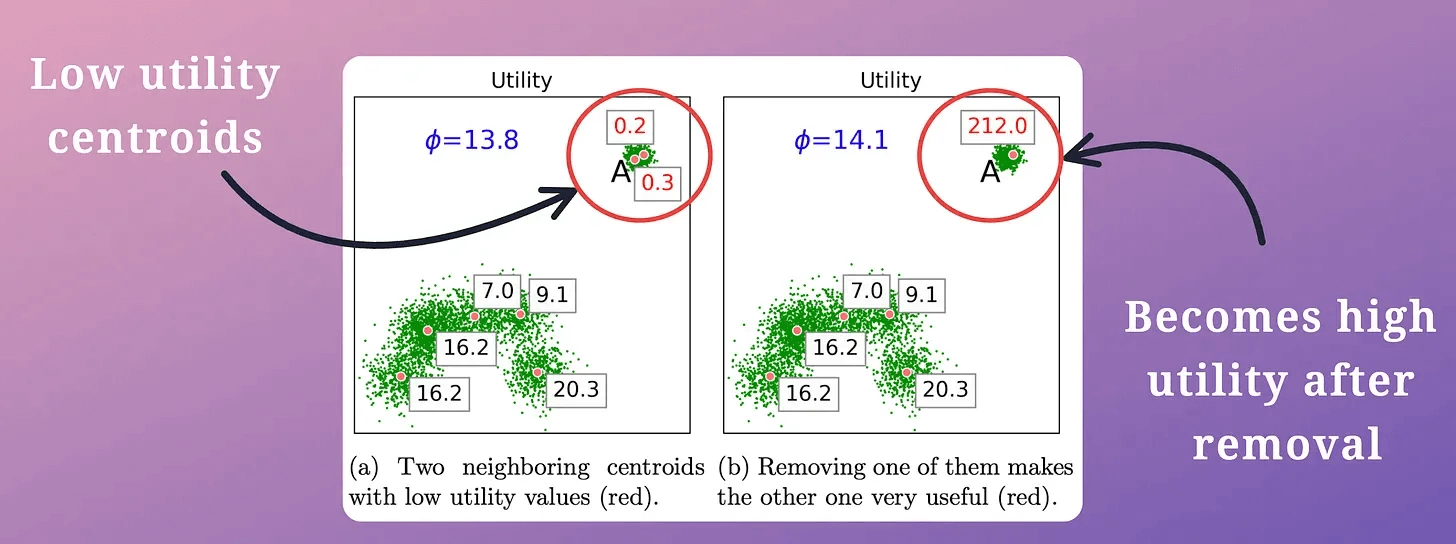
This is determined using the “utility” of a centroid.
A centroid’s utility is proportional to its distance from all other centroids.
The greater the distance, the more isolated it will be; hence, the more the utility.
In other words, if two centroids are close, they may lie in the same cluster.
Thus, we must remove one of them, as demonstrated below (in the top right cluster):
This is repeated until all “m” low-utility centroids have been removed.
This gives back “k” centroids.
Finally, we run KMeans with these “k” centroids only once.
Step 4: Decrease m by 1.
Step 5: Repeat Steps 2 to 4 until m=0.
Done!
Why does Breathing Kmeans work?
These repeated breathing cycles (breathe-in and breathe-out steps) almost always provide a faster and better solution than standard KMeans with repetitions.
In each cycle:
New centroids are added at “good” locations. This helps in splitting clusters occupied by a single centroid.
Low-utility centroids are removed. This helps eliminate centroids that are likely in the same cluster.
As a result, it is expected to converge to the optimal solution faster.
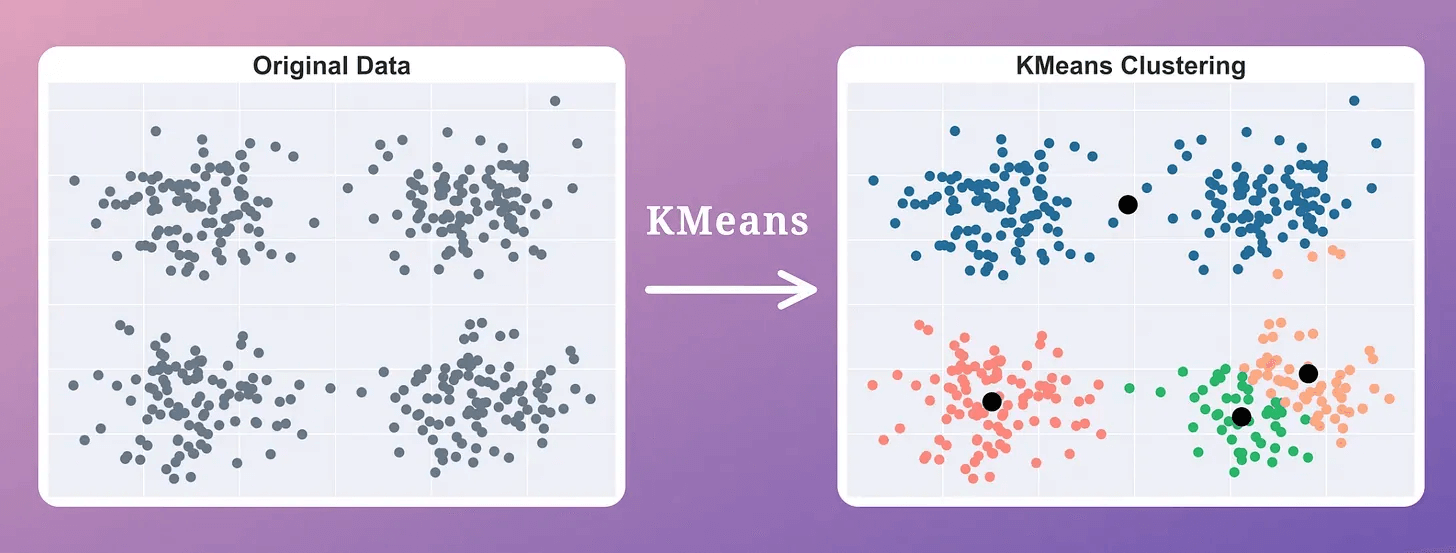
The effectiveness of Breathing KMeans over KMeans is evident from the image below:
KMeans produced two misplaced centroids.
Breathing KMeans accurately clustered the data with a 50% run-time improvement.
Isn’t that a significant upgrade to KMeans?
That said, data conformity is another big issue with KMeans, which makes it highly inapplicable in many data situations.
These three guides cover distribution-based and density-based clustering, which address KMeans’ limitations in specific data situations:
Gaussian Mixture Models (GMMs) [derived and implemented from scratch using NumPy].
DBSCAN++: The Faster and Scalable Alternative to DBSCAN Clustering [with implementation].
HDBSCAN—An Algorithmic Deep Dive [with implementation].
👉 Over to you: What are some other ways to improve KMeans’ clustering and its run-time?
Thanks for reading!