MissForest: A Better Alternative To Zero (or Mean) Imputation
Missing value imputation using Random Forest.
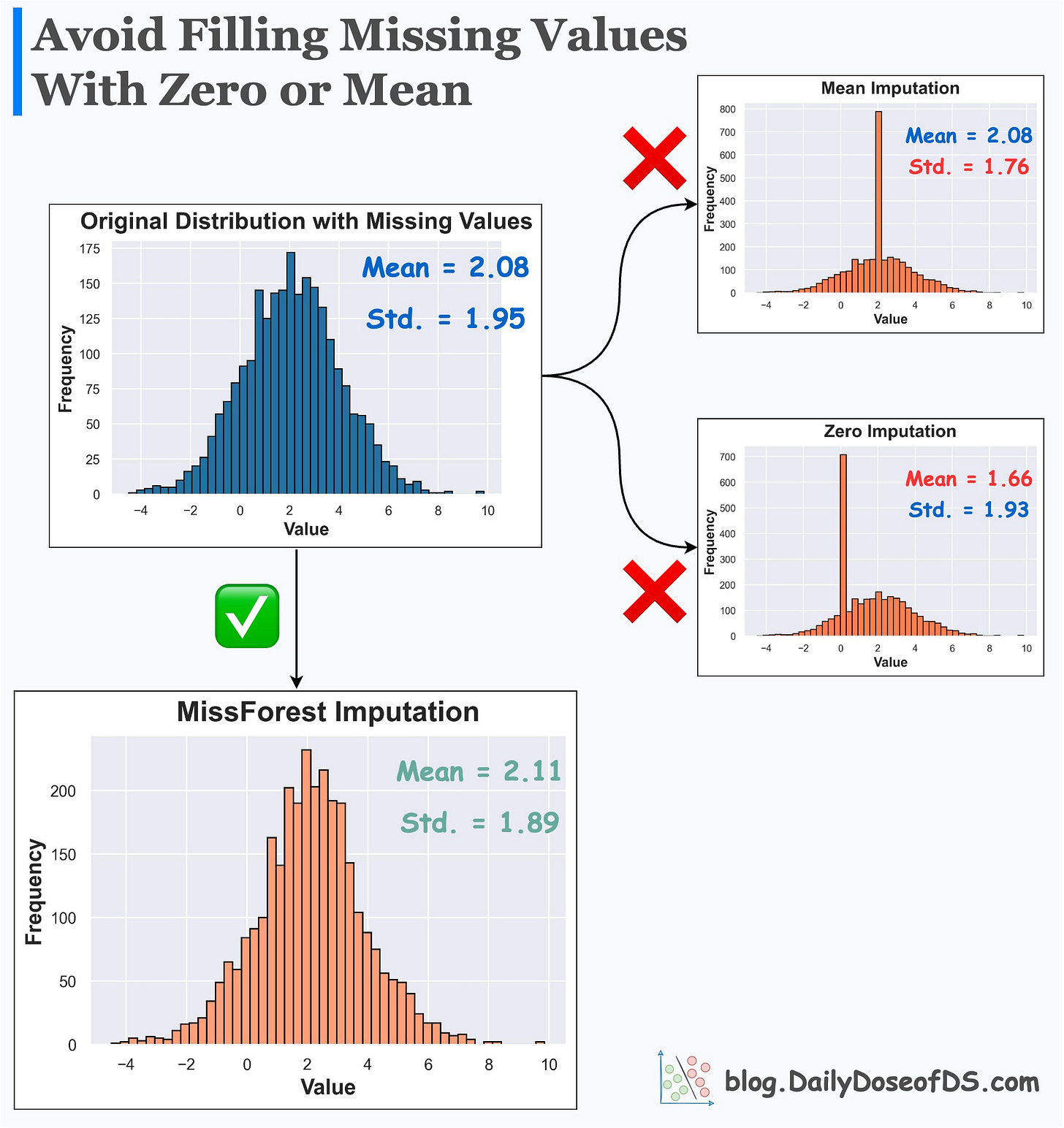
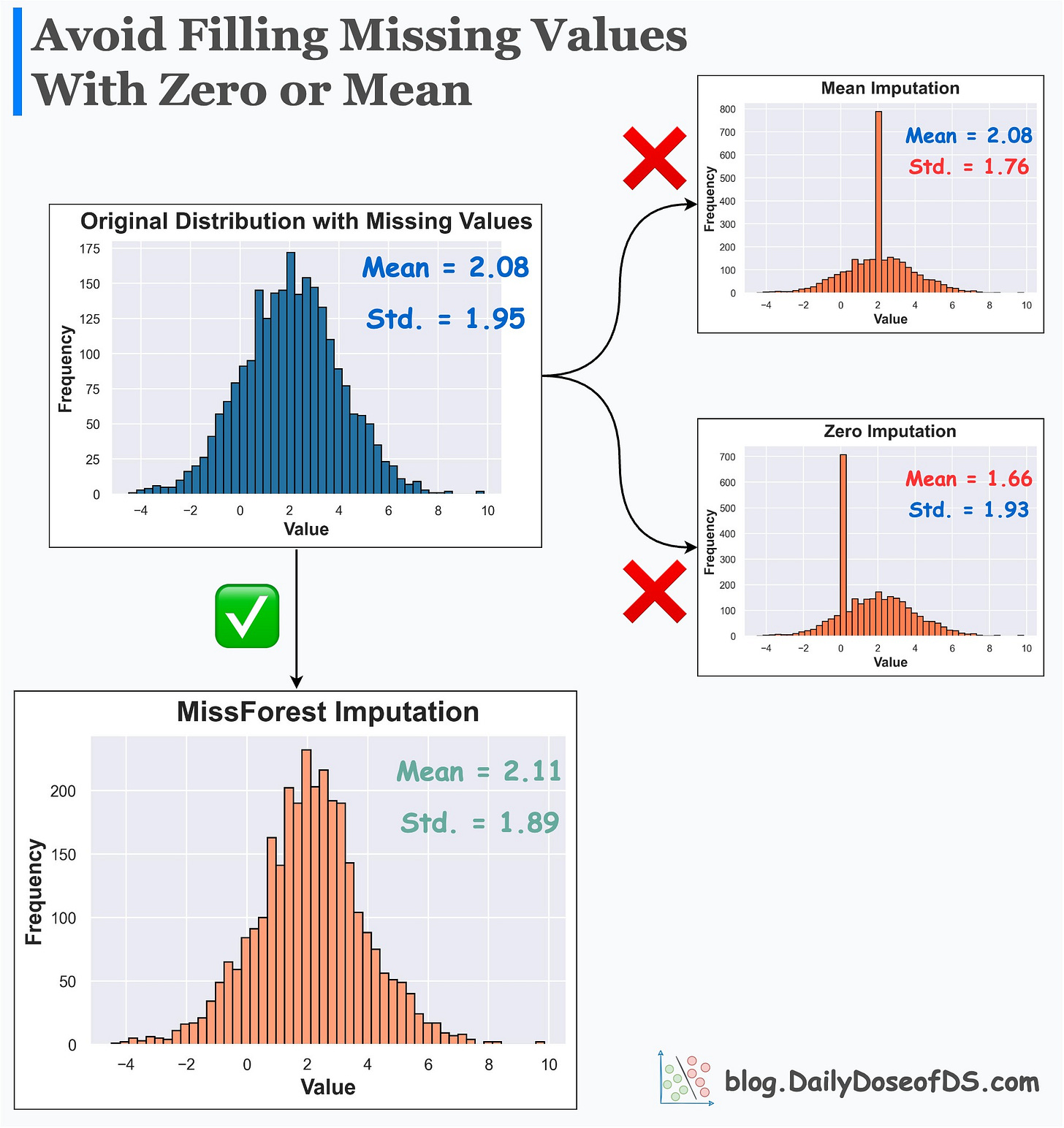
Replacing (imputing) missing values with mean or zero or any other fixed value:
alters summary statistics
changes the distribution
inflates the presence of a specific value
This can lead to:
inaccurate modeling
incorrect conclusions, and more.
Instead, always try to impute missing values with more precision.
In one of the earlier posts, we discussed kNN imputer. Today’s post builds on that by addressing its limitations, which are:
High run-time for imputation — especially for high-dimensional datasets.
Issues with distance calculation in case of categorical non-missing features.
Requires feature scaling, etc.
MissForest imputer is another reliable choice for missing value imputation when your data is missing at random (MAR).
As the name suggests, it imputes missing values using the Random Forest algorithm.
The following figure depicts how it works:
Step 1: To begin, impute the missing feature with a random guess — Mean, Median, etc.
Step 2: Model the missing feature using Random Forest.
Step 3: Impute ONLY originally missing values using Random Forest’s prediction.
Step 4: Back to Step 2. Use the imputed dataset from Step 3 to train the next Random Forest model.
Step 5: Repeat until convergence (or max iterations).
In case of multiple missing features, the idea (somewhat) stays the same:
Impute features sequentially in increasing order missingness — features with fewer missing values are imputed first.
Its effectiveness over Mean/Zero imputation is evident from the image below.
Mean/Zero alters the summary statistics and distribution.
MissForest imputer preserves them.
What’s more, MissForest can impute even if the data has categorical non-missing features.
MissForest is based on Random Forest, so one can impute from categorical and continuous data.
Get started with MissForest imputer: MissingPy MissForest.
👉 Over to you: What are some other better ways to impute missing values?
👉 If you liked this post, don’t forget to leave a like ❤️. It helps more people discover this newsletter on Substack and tells me that you appreciate reading these daily insights.
The button is located towards the bottom of this email.
Thanks for reading!
Latest full articles
If you’re not a full subscriber, here’s what you missed last month:
DBSCAN++: The Faster and Scalable Alternative to DBSCAN Clustering
Federated Learning: A Critical Step Towards Privacy-Preserving Machine Learning
You Cannot Build Large Data Projects Until You Learn Data Version Control!
Sklearn Models are Not Deployment Friendly! Supercharge Them With Tensor Computations.
Deploy, Version Control, and Manage ML Models Right From Your Jupyter Notebook with Modelbit
Gaussian Mixture Models (GMMs): The Flexible Twin of KMeans.
To receive all full articles and support the Daily Dose of Data Science, consider subscribing:
👉 Tell the world what makes this newsletter special for you by leaving a review here :)
👉 If you love reading this newsletter, feel free to share it with friends!





Better still is not to impute anything but rather to leave it up to each model's perspective how to treat missing values. For example "distance functions" can be customised and in some cases asymmetric (e.g. reflecting some aspect of the application domain). Preprocessing data presupposes downstream purposes (that might change over time).
Outlier-tolerant e.g. median would be better than mean - though in your example that would just place the spike in a "better" place.