MissForest and kNN Imputation for Data Missing at Random
...with a practical demonstration.
The imputation strategy for missing data largely depends on the type of missingness, which can be of three types:
Missing completely at random (MCAR): Data is genuinely missing at random and has no relation to any observed or unobserved variables.
Missing at random (MAR): The missingness of one feature can be explained by other observed features in the dataset.
Missing not at random (MNAR): Missingness is either attributed to the missing value itself or the feature(s) that we didn’t collect data for.
I have observed missing at random (MAR) appearing relatively much more in practical situations than the other two, so today, I want to share two imputation strategies I typically prefer to use in such a case.
I have provided the jupyter notebook for today’s issue towards the end.
Let’s begin!
#1) kNN imputation
As the name suggests, it imputes missing values using the k-nearest neighbors algorithm.
More specifically, missing features are imputed by running a kNN on non-missing feature values.
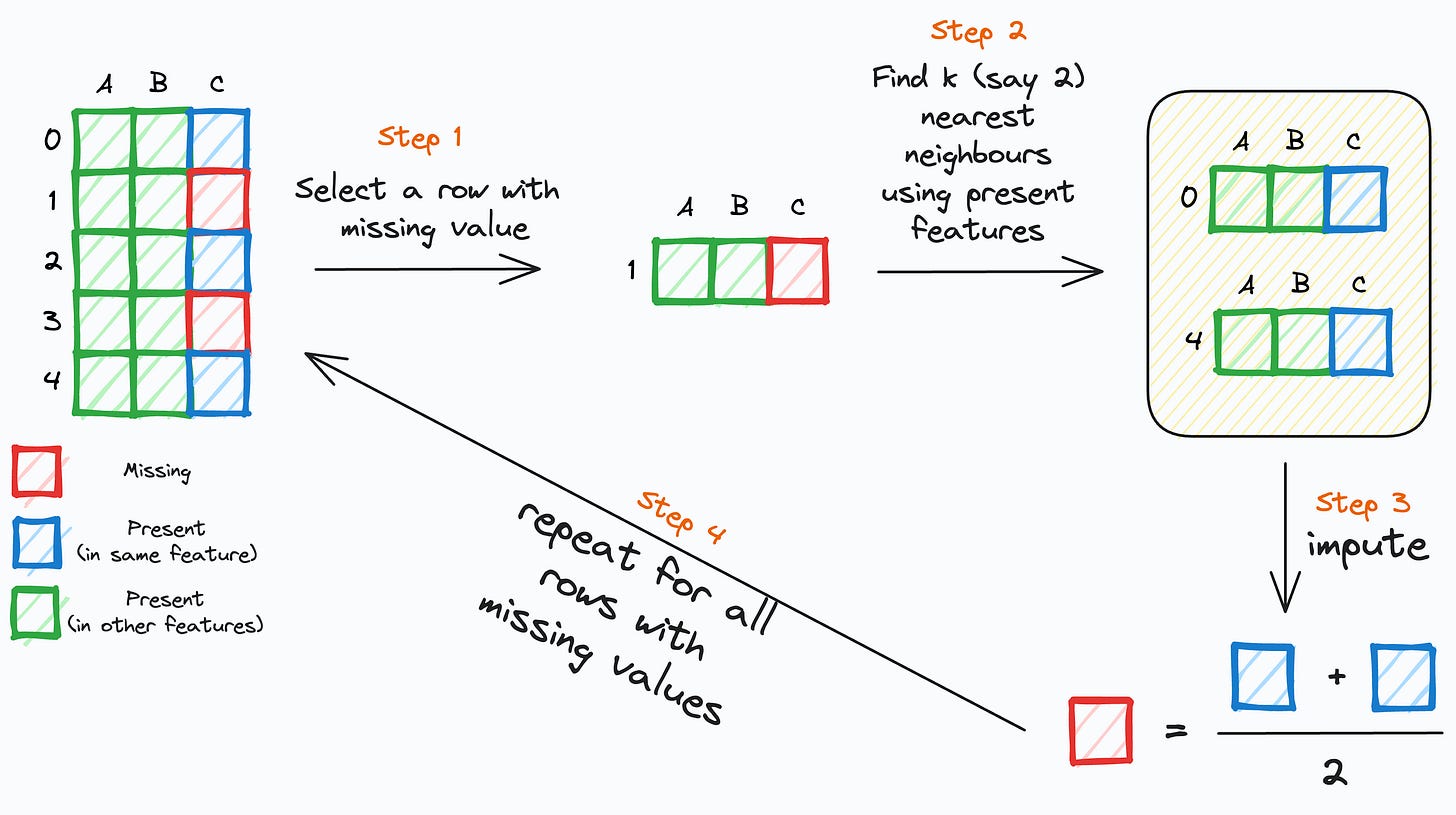
The following image depicts how it works:
Step 1: Select a row (
r) with a missing value.Step 2: Find its
knearest neighbors using the non-missing feature values.Step 3: Impute the missing feature of the row (
r) using the corresponding non-missing values ofknearest neighbor rows.Step 4: Repeat for all rows with missing values.
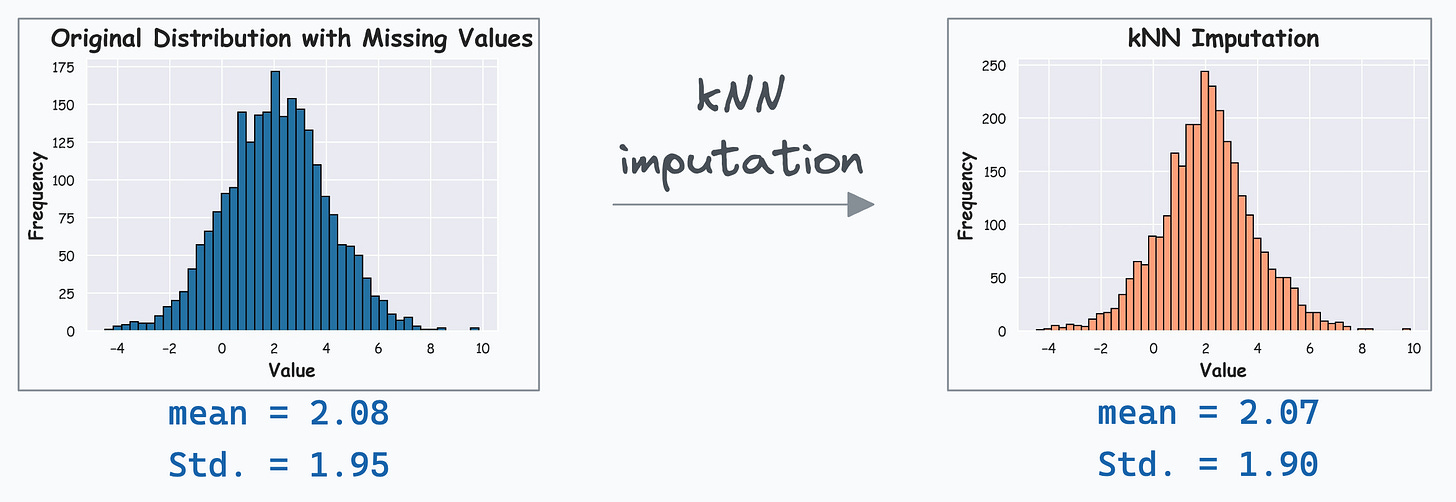
Its effectiveness over mean/zero imputation is evident from the demo below.
On the left, we have a feature distribution with missing values. Assuming we have already validated that the data was missing at random (MAR), using mean/zero alters the summary statistics and distribution, as depicted below.
However, as depicted below, kNN imputer appears to be more reliable, and it preserves the summary statistics:
#2) MissForest
Some major issues with kNN imputation are:
It has a high run-time for imputation — especially for high-dimensional datasets.
It raises issues with distance calculation in the case of categorical non-missing features.
It requires feature scaling, etc.
MissForest imputer is another reliable choice for missing value imputation when data is missing at random (MAR).
As the name suggests, it imputes missing values using the Random Forest algorithm.
The following figure depicts how it works:
Step 1: To begin, impute the missing feature with a random guess — Mean, Median, etc.
Step 2: Model the missing feature using Random Forest.
Step 3: Impute ONLY originally missing values using Random Forest’s prediction.
Step 4: Back to Step 2. Use the imputed dataset from Step 3 to train the next Random Forest model.
Step 5: Repeat until convergence (or max iterations).
In case of multiple missing features, the idea (somewhat) stays the same:
Impute features sequentially in increasing order missingness — features with fewer missing values are imputed first.
Its effectiveness over mean/zero imputation is evident from the demo below.
Like before, on the left, we have a feature distribution with missing values. Again, assuming we have already validated that the data was missing at random (MAR), using mean/zero alters the summary statistics and distribution, as depicted below.
However, as depicted below, MissForest appears more reliable and preserves the summary statistics.
Get started with kNN imputation and MissForest by downloading this Jupyter notebook: kNN imputation and MissForest notebook.
👉 Over to you: What are some other better ways to impute missing values when data is missing at random (MAR)?
Are you overwhelmed with the amount of information in ML/DS?
Every week, I publish no-fluff deep dives on topics that truly matter to your skills for ML/DS roles.
For instance:
A Beginner-friendly Introduction to Kolmogorov Arnold Networks (KANs).
5 Must-Know Ways to Test ML Models in Production (Implementation Included).
Understanding LoRA-derived Techniques for Optimal LLM Fine-tuning
8 Fatal (Yet Non-obvious) Pitfalls and Cautionary Measures in Data Science
Implementing Parallelized CUDA Programs From Scratch Using CUDA Programming
You Are Probably Building Inconsistent Classification Models Without Even Realizing.
How To (Immensely) Optimize Your Machine Learning Development and Operations with MLflow.
And many many more.
Join below to unlock all full articles:
SPONSOR US
Get your product in front of 78,000 data scientists and other tech professionals.
Our newsletter puts your products and services directly in front of an audience that matters — thousands of leaders, senior data scientists, machine learning engineers, data analysts, etc., who have influence over significant tech decisions and big purchases.
To ensure your product reaches this influential audience, reserve your space here or reply to this email to ensure your product reaches this influential audience.










Thanks for the newsletter, although quite impressive, this seems like a lot of overhead for imputing missing values compared to just imputing using the mean. I guess when it is a high stakes case and the missing values are few, this makes sense. Would love to get your comments on the kind of data that where it would necessitate this or where this kind of imputing works well. Enjoyed the read
Does this imputation apply to categorical features?