Popular Interview Question: PCA vs. t-SNE
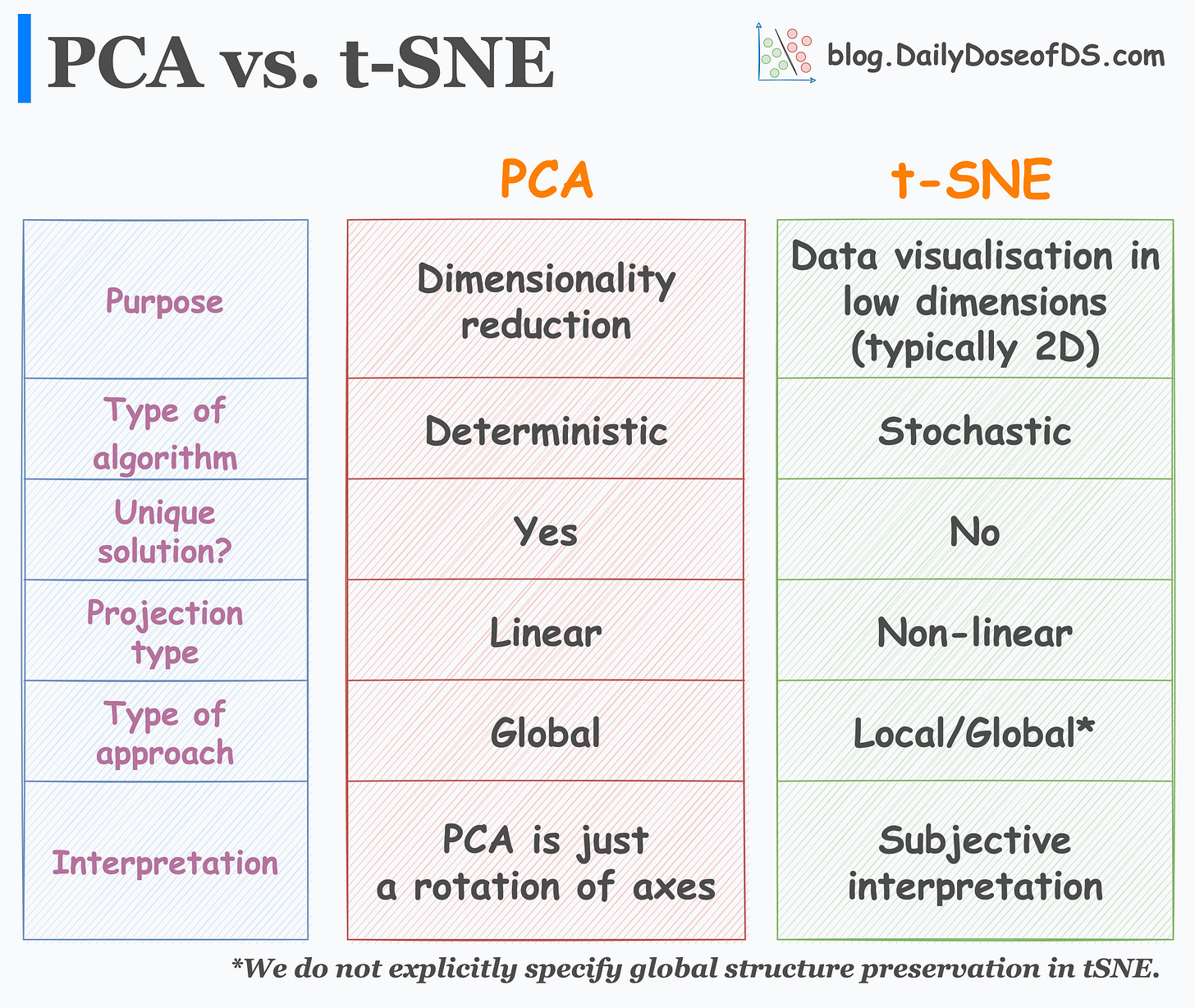
Comparing both algorithms on six parameters.
Many people face difficulties in understanding the precise differences between PCA and t-SNE algorithms.
If you are one of them, I prepared the following table for you which neatly summarizes the major differences between the two algorithms:
Learn more about their motivation, full mathematics, custom implementations (without sklearn), limitations, etc., by reading these two in-depth articles:
Let’s understand their differences:
#1) Purpose
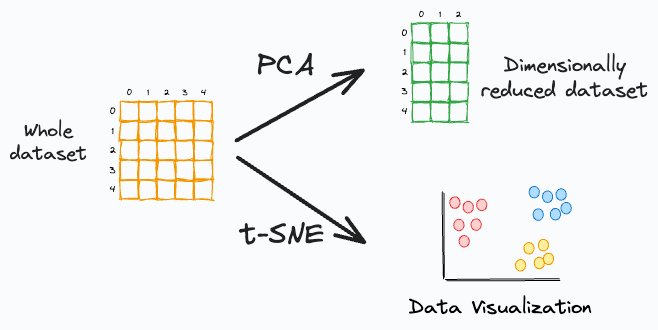
While many interpret PCA as a data visualization algorithm, it is primarily a dimensionality reduction algorithm.
t-SNE, however, is a data visualization algorithm. We use it to project high-dimensional data to low dimensions (primarily 2D).
#2) Type of algorithm
PCA is a deterministic algorithm. Thus, if we run the algorithm twice on the same dataset, we will ALWAYS get the same result.
t-SNE, however, is a stochastic algorithm. Thus, rerunning the algorithm can lead to entirely different results. Can you explain why? Share your answers :)
#3) Uniqueness of solution
As far as uniqueness and interpretation of results is concerned…
PCA always has a unique solution for the projection of data points. Simply put, PCA is just a rotation of axes such that the new features we get are uncorrelated.
t-SNE, as discussed above, can provide entirely different results, and its interpretation is subjective in nature.
#4) Projection type
PCA is a linear dimensionality reduction approach. It can only find a linear subspace to project the given dataset. KernelPCA addresses this, which we covered in detail here: KernelPCA vs. PCA for Dimensionality Reduction.
t-SNE is a non-linear approach. It can handle non-linear datasets.
#5) Projection approach
PCA only aims to retain the global variance of the data. Thus, local relationships (such as clusters) are often lost after projection, as shown below:
t-SNE preserves local relationships. Thus, data points in a cluster in the high-dimensional space are much more likely to lie together in the low-dimensional space.
In t-SNE, we do not explicitly specify global structure preservation. But it typically does create well-separated clusters.
Nonetheless, it is important to note that the distance between two clusters in low-dimensional space is NEVER an indicator of cluster separation in high-dimensional space. I wrote a cautionary guide on t-SNE here: How To Avoid Getting Misled by t-SNE Projections?
If you are interested in learning more about their motivation, full mathematics, custom implementations, limitations, etc., read these two in-depth articles:
👉 Over to you: What other differences between t-SNE and PCA did I miss?
Thanks for reading!
Whenever you are ready, here’s one more way I can help you:
Every week, I publish 1-2 in-depth deep dives (typically 20+ mins long). Here are some of the latest ones that you will surely like:
[FREE] A Beginner-friendly and Comprehensive Deep Dive on Vector Databases.
A Detailed and Beginner-Friendly Introduction to PyTorch Lightning: The Supercharged PyTorch
You Are Probably Building Inconsistent Classification Models Without Even Realizing
PyTorch Models Are Not Deployment-Friendly! Supercharge Them With TorchScript.
Federated Learning: A Critical Step Towards Privacy-Preserving Machine Learning.
You Cannot Build Large Data Projects Until You Learn Data Version Control!
To receive all full articles and support the Daily Dose of Data Science, consider subscribing:
👉 If you love reading this newsletter, share it with friends!
👉 Tell the world what makes this newsletter special for you by leaving a review here :)