The Limitations of DBSCAN Clustering Which Many Often Overlook
...And here's a better alternative to work with.
DBSCAN is a density-based clustering, which clusters data points based on density.
This makes it more robust than algorithms like KMeans because:
Being “density-based”, it can identify clusters of varying shapes.
While KMeans can only create globular clusters.
A comparison between DBSCAN and KMeans is shown below:
KMeans attempts to form globular clusters. Hence, it fails to identify correct clusters.
DBSCAN relies on the concept of “density”, making it more robust.
Yet, it is important to note that DBSCAN also has some limitations, which many often overlook.
Let’s understand these today.
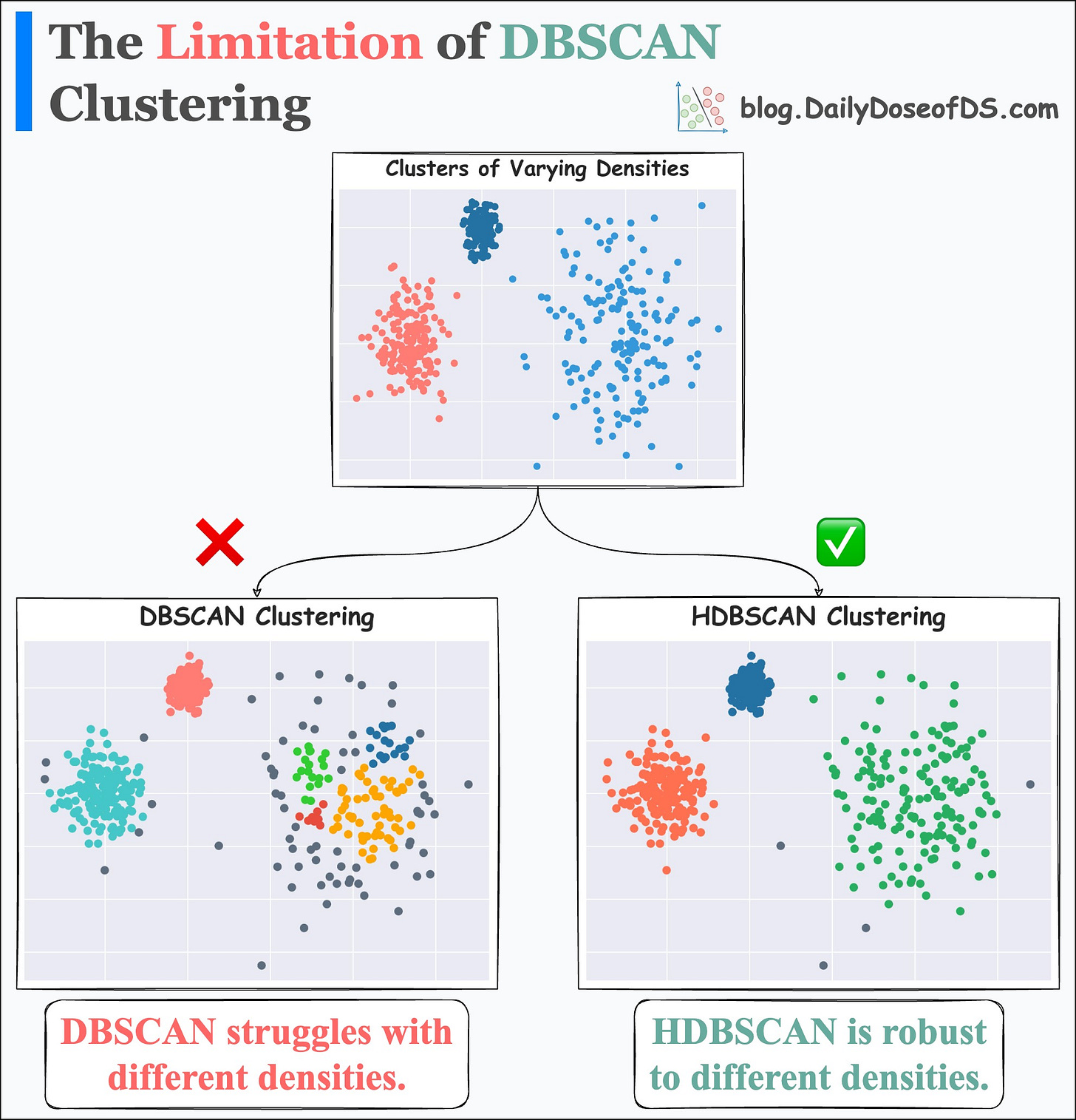
To begin, DBSCAN assumes that the local density of data points is (somewhat) globally uniform. This is governed by its eps parameter.
Thus, it may struggle to identify clusters with varying densities.
This may need several hyperparameter tuning attempts to get promising results.
HDBSCAN can be a better choice for density-based clustering.
It relaxes the assumption of local uniform density, which makes it more robust to clusters of varying densities by exploring many different density scales.
Its effectiveness is evident from the image below:
On a dataset with three clusters, each with varying densities:
DBSCAN struggles to identify correct clusters.
HDBSCAN is found to be more robust.
What's more:
DBSCAN is a scale variant algorithm. Thus, clustering results for data X, 2X, 3X, etc., can be entirely different.
On the other hand, HDBSCAN is scale-invariant. So, clustering results remain the same across different scales of data.
We can also verify this experimentally:
The results vary with the data scale for DBSCAN.
The results remain unaltered with the scale for HDBSCAN.
👉 Over to you: Can you explain why HDBSCAN is scale-invariant?
👉 If you liked this post, don’t forget to leave a like ❤️. It helps more people discover this newsletter on Substack and tells me that you appreciate reading these daily insights. The button is located towards the bottom of this email.
Thanks for reading!
In case you missed it
Yesterday, I introduced The Daily Dose of Data Science Lab, a cohort-based platform for you to:
Attend weekly live sessions (office hours) hosted by me and invited guests.
Enroll in self-paced and live courses.
Get private mentoring.
Join query discussions.
Find answers to your data-related problems.
Refer to the internal data science resources, and more.
To ensure an optimal and engaging experience, The Lab will always operate at a capacity of 120 active participants.
So, if you are interested in receiving further updates about The Lab, please fill out this form: The Lab interest form.
Note: Filling out the form DOES NOT mean you must join The Lab. This is just an interest form to indicate that you are interested in learning more before making a decision.
I will be sharing more details with the respondents soon.
Thank you :)
Latest full articles
If you’re not a full subscriber, here’s what you missed last month:
Deploy, Version Control, and Manage ML Models Right From Your Jupyter Notebook with Modelbit
Model Compression: A Critical Step Towards Efficient Machine Learning.
Generalized Linear Models (GLMs): The Supercharged Linear Regression.
Gaussian Mixture Models (GMMs): The Flexible Twin of KMeans.
To receive all full articles and support the Daily Dose of Data Science, consider subscribing:
👉 Tell the world what makes this newsletter special for you by leaving a review here :)
👉 If you love reading this newsletter, feel free to share it with friends!






Thanks for this crisp and informative post Avi!