The Ultimate Comparison Between PCA and t-SNE Algorithm
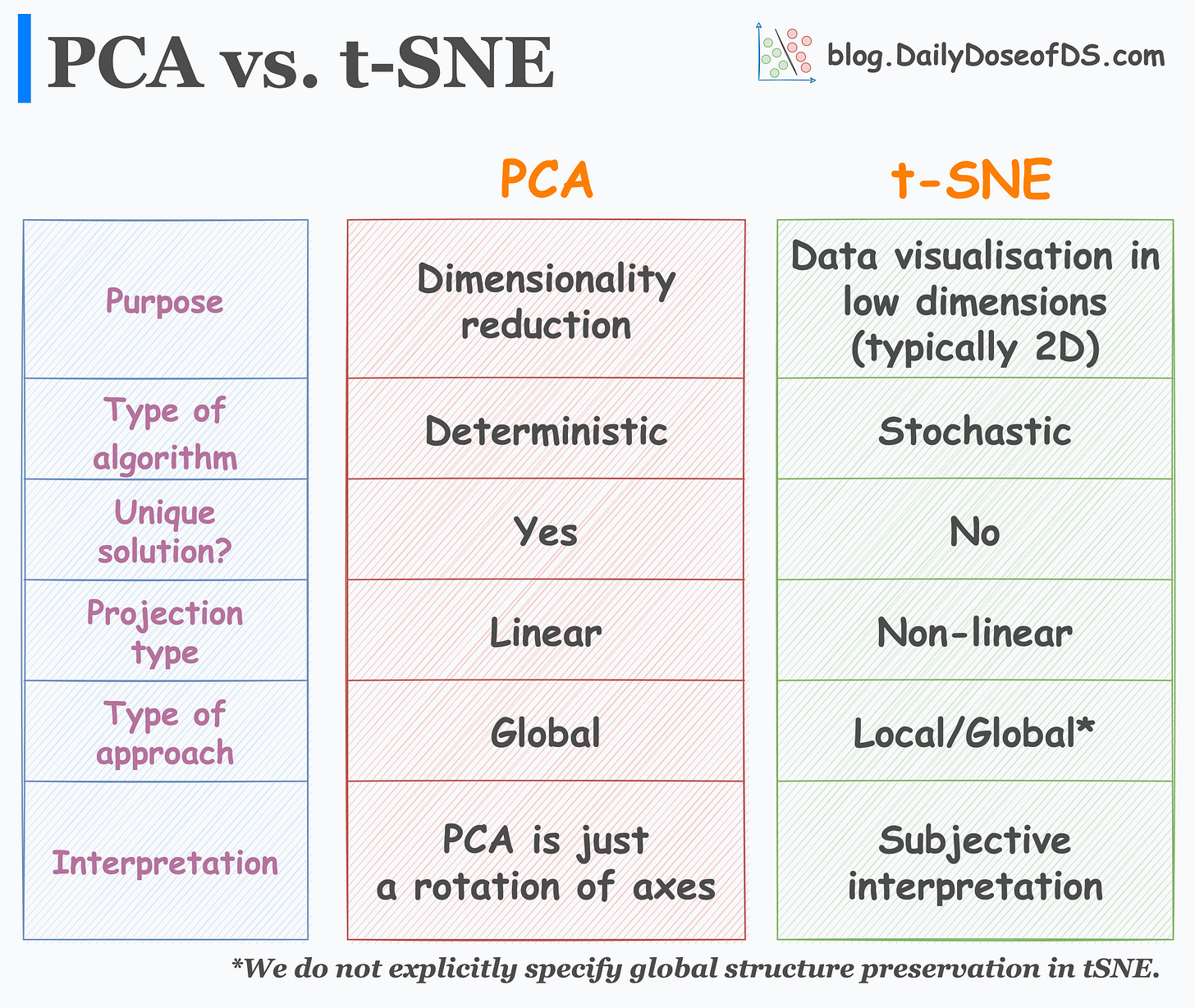
Comparing both algorithms on six parameters.
In earlier newsletter issues, we discussed PCA and t-SNE individually.
Yet, a formal comparison between the two approaches is still left to be covered.
Let’s do it today.
The visual below neatly summarizes the major differences between the two algorithms:
First and foremost, let’s understand their purpose:
While many interpret PCA as a data visualization algorithm, it is primarily a dimensionality reduction algorithm.
t-SNE, however, is a data visualization algorithm. We use it to project high-dimensional data to low dimensions (primarily 2D).
Moving on:
PCA is a deterministic algorithm. Thus, if we run the algorithm twice on the same dataset, we will ALWAYS get the same result.
t-SNE, however, is a stochastic algorithm. Thus, rerunning the algorithm can lead to entirely different results. Can you explain why? Share your answers :)
As far as uniqueness and interpretation of results is concerned:
PCA always has a unique solution for the projection of data points. Simply put, PCA is just a rotation of axes such that the new features we get are uncorrelated.
t-SNE, as discussed above, can provide entirely different results, and its interpretation is subjective in nature.
Next, how do they project data?
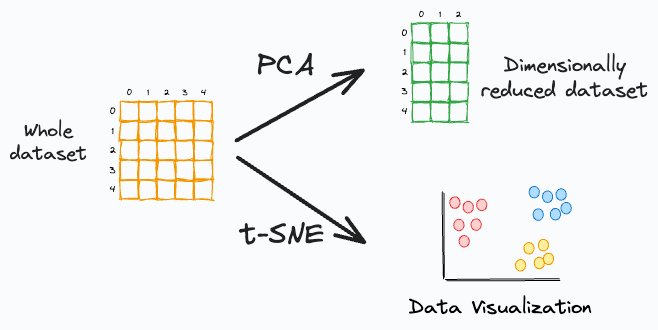
PCA is a linear dimensionality reduction approach. Thus, it is not well-suited if we have a non-linear dataset (which is often true), as shown below:
t-SNE is a non-linear approach. It can handle non-linear datasets.
During dimensionality reduction:
PCA only aims to retain the global variance of the data. Thus, local relationships (such as clusters) are often lost after projection, as shown below:
t-SNE preserves local relationships. Thus, data points in a cluster in the high-dimensional space are much more likely to lie together in the low-dimensional space.
In t-SNE, we do not explicitly specify global structure preservation. But it typically does create well-separated clusters.
Nonetheless, it is important to note that the distance between two clusters in low-dimensional space is NEVER an indicator of cluster separation in high-dimensional space.
If you are interested in learning more about their motivation, mathematics, custom implementations, limitations, etc., feel free to read these two in-depth articles:
👉 Over to you: What other differences between t-SNE and PCA did I miss?
👉 If you liked this post, don’t forget to leave a like ❤️. It helps more people discover this newsletter on Substack and tells me that you appreciate reading these daily insights. The button is located towards the bottom of this email.
Thanks for reading!
In case you missed it
Recently, I introduced The Daily Dose of Data Science Lab, a cohort-based platform for you to:
Attend weekly live sessions (office hours) hosted by me and invited guests.
Enroll in self-paced and live courses.
Get private mentoring.
Join query discussions.
Find answers to your data-related problems.
Refer to the internal data science resources, and more.
To ensure an optimal and engaging experience, The Lab will always operate at a capacity of 120 active participants.
So, if you are interested in receiving further updates about The Lab, please fill out this form: The Lab interest form.
Note: Filling out the form DOES NOT mean you must join The Lab. This is just an interest form to indicate that you are interested in learning more before making a decision.
I will be sharing more details with the respondents soon.
Thank you :)
Latest full articles
If you’re not a full subscriber, here’s what you missed last month:
Deploy, Version Control, and Manage ML Models Right From Your Jupyter Notebook with Modelbit
Model Compression: A Critical Step Towards Efficient Machine Learning.
Generalized Linear Models (GLMs): The Supercharged Linear Regression.
Gaussian Mixture Models (GMMs): The Flexible Twin of KMeans.
To receive all full articles and support the Daily Dose of Data Science, consider subscribing:
👉 Tell the world what makes this newsletter special for you by leaving a review here :)
👉 If you love reading this newsletter, feel free to share it with friends!






It would be nice to see some comparison with UMAP