HDBSCAN vs. DBSCAN
What limitations does HDBSCAN address?
We have discussed DBSCAN and its scalable alternative, DBSCAN++, in this newsletter before: DBSCAN and DBSCAN++.
Today, I want to dive into HDBSCAN and share how it differs from DBSCAN.
Why DBSCAN?
While KMeans is widely used due to its simplicity and effectiveness as a clustering algorithm, it has many limitations:
It does not account for cluster covariance.
It can only produce spherical clusters. As shown below, even if the data has non-circular clusters, it still produces round clusters.
Density-based algorithms, like DBSCAN, quite effectively address these limitations, as depicted below:
On a side note, if you have no idea about DBSCAN, I would highly recommend watching this animated video I published in this newsletter some time back:
Coming back to the topic…
But, of course, like any other algorithm, DBSCAN also has some limitations.
To begin, DBSCAN assumes that the local density of data points is (somewhat) globally uniform. This is governed by its eps parameter.
Thus, it may struggle to identify clusters with varying densities. This may need several hyperparameter tuning attempts to get promising results.
HDBSCAN can be a better choice for density-based clustering.
It relaxes the assumption of local uniform density, which makes it more robust to clusters of varying densities by exploring many different density scales.
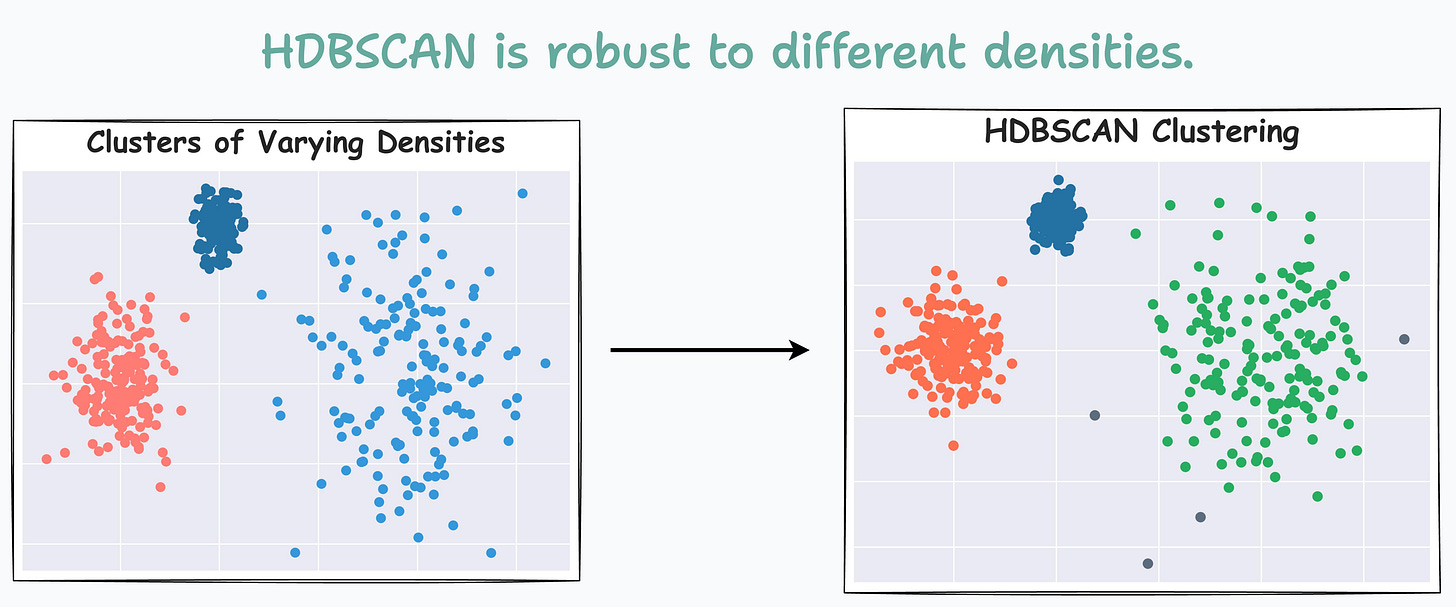
For instance, consider the clustering results obtained with DBSCAN on the dummy dataset below, where each cluster has different densities:
It is clear that DBSCAN produces bad clustering results.
Now compare it with HDBSCAN results depicted below:
On a dataset with three clusters, each with varying densities, HDBSCAN is found to be more robust.
There’s one more thing I love about HDBSCAN:
DBSCAN is a scale variant algorithm. Thus, clustering results for data X, 2X, 3X, etc., can be entirely different.
On the other hand, HDBSCAN is scale-invariant. So, clustering results remain the same across different scales of data.
This is depicted below:
We have DBSCAN on the left, and we can see that the results vary with the scale of the data.
However, clustering from HDBSCAN (on the right) remains unaltered with the scale for HDBSCAN.
Isn’t that cool?
I have been wanting to write an extensive article about HDBSCAN for some time now. Let me know if you are interested in learning more about it:
👉 Over to you: Can you explain why HDBSCAN is scale-invariant?
Are you overwhelmed with the amount of information in ML/DS?
Every week, I publish no-fluff deep dives on topics that truly matter to your skills for ML/DS roles.
For instance:
A Beginner-friendly Introduction to Kolmogorov Arnold Networks (KANs).
5 Must-Know Ways to Test ML Models in Production (Implementation Included).
Understanding LoRA-derived Techniques for Optimal LLM Fine-tuning
8 Fatal (Yet Non-obvious) Pitfalls and Cautionary Measures in Data Science
Implementing Parallelized CUDA Programs From Scratch Using CUDA Programming
You Are Probably Building Inconsistent Classification Models Without Even Realizing.
And many many more.
Join below to unlock all full articles:
SPONSOR US
Get your product in front of 80,000 data scientists and other tech professionals.
Our newsletter puts your products and services directly in front of an audience that matters — thousands of leaders, senior data scientists, machine learning engineers, data analysts, etc., who have influence over significant tech decisions and big purchases.
To ensure your product reaches this influential audience, reserve your space here or reply to this email to ensure your product reaches this influential audience.









Insightful. Always useful to know the limitations of an algorithm from the PoV of input data
HDBSCAN is not actually scale invariant. It is very robust to scale changes, but technically there exist scale changes that will result in different clusterings for almost any dataset that has multiple clusters.